Abstract
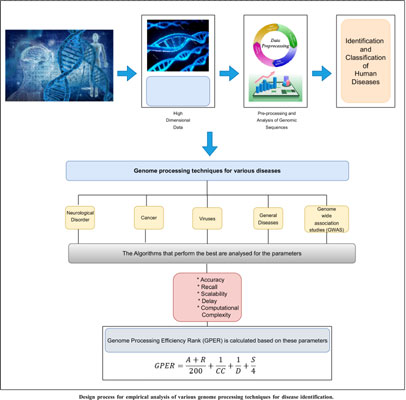
Human gene sequences are considered a primary source of comprehensive information about different body conditions. A wide variety of diseases including cancer, heart issues, brain issues, genetic issues, etc. can be pre-empted via efficient analysis of genomic sequences. Researchers have proposed different configurations of machine learning models for processing genomic sequences, and each of these models varies in terms of their performance & applicability characteristics. Models that use bioinspired optimizations are generally slower, but have superior incrementalperformance, while models that use one-shot learning achieve higher instantaneous accuracy but cannot be scaled for larger disease-sets. Due to such variations, it is difficult for genomic system designers to identify optimum models for their application-specific & performance-specific use cases. To overcome this issue, a detailed survey of different genomic processing models in terms of their functional nuances, application-specific advantages, deployment-specific limitations, and contextual future scopes is discussed in this text. Based on this discussion, researchers will be able to identify optimal models for their functional use cases. This text also compares the reviewed models in terms of their quantitative parameter sets, which include, the accuracy of classification, delay needed to classify large-length sequences, precision levels, scalability levels, and deployment cost, which will assist readers in selecting deployment-specific models for their contextual clinical scenarios. This text also evaluates a novel Genome Processing Efficiency Rank (GPER) for each of these models, which will allow readers to identify models with higher performance and low overheads under real-time scenarios.
Keywords: Genome processing, machine learning, disease, deep learning, gene network, classification.
[http://dx.doi.org/10.1109/TCBB.2018.2834529]
[http://dx.doi.org/10.1109/TCBB.2020.2993301]
[http://dx.doi.org/10.1109/JBHI.2018.2870728] [PMID: 31283472]
[http://dx.doi.org/10.1109/TCBB.2017.2774802]
[http://dx.doi.org/10.1109/TCBB.2018.2817624]
[http://dx.doi.org/10.1109/ACCESS.2019.2941955]
[http://dx.doi.org/10.1109/TIM.2021.3107056]
[http://dx.doi.org/10.1109/ACCESS.2019.2935515]
[http://dx.doi.org/10.1109/TCBB.2018.2835459]
[http://dx.doi.org/10.1109/ACCESS.2020.3020592]
[http://dx.doi.org/10.1109/ACCESS.2019.2962091]
[http://dx.doi.org/10.1109/TCBB.2020.3027392]
[http://dx.doi.org/10.1109/JBHI.2021.3106787]
[http://dx.doi.org/10.1109/ACCESS.2019.2954994]
[http://dx.doi.org/10.1109/TCBB.2019.2938512]
[http://dx.doi.org/10.1109/ACCESS.2021.3065280]
[http://dx.doi.org/10.3390/life12020279] [PMID: 35207566]
[http://dx.doi.org/10.2174/1389202923666220527114108] [PMID: 36777878]
[PMID: 31680165]
[http://dx.doi.org/10.2174/1389202920666190326145459] [PMID: 31929727]
[http://dx.doi.org/10.1109/ACCESS.2020.2975585]
[http://dx.doi.org/10.1109/TCBB.2018.2814043]
[http://dx.doi.org/10.1109/ACCESS.2020.2987071]
[http://dx.doi.org/10.1109/TCBB.2019.2949039]
[http://dx.doi.org/10.1016/j.artmed.2022.102276]
[http://dx.doi.org/10.1109/JBHI.2020.2987034] [PMID: 32287029]
[http://dx.doi.org/10.26599/BDMA.2020.9020025]
[http://dx.doi.org/10.1109/TETCI.2020.3014923]
[http://dx.doi.org/10.1109/TCBB.2012.80]
[http://dx.doi.org/10.1109/ACCESS.2022.3186998]
[http://dx.doi.org/10.1109/ACCESS.2020.3013666]
[http://dx.doi.org/10.1109/ACCESS.2017.2778268]
[http://dx.doi.org/10.1109/TBCAS.2015.2487299] [PMID: 26841412]
[http://dx.doi.org/10.1109/TFUZZ.2019.2914629]
[http://dx.doi.org/10.1109/ACCESS.2019.2923687]
[http://dx.doi.org/10.1109/TCBB.2020.2994649]
[http://dx.doi.org/10.2174/1389202923666220927105311] [PMID: 36778194]
[http://dx.doi.org/10.1109/ACCESS.2021.3073728] [PMID: 34812391]
[http://dx.doi.org/10.1109/TCBB.2013.141]
[http://dx.doi.org/10.1109/ACCESS.2020.3031387] [PMID: 34976561]
[http://dx.doi.org/10.1109/TCBB.2019.2897683]
[http://dx.doi.org/10.1109/ACCESS.2020.3002923]
[http://dx.doi.org/10.1109/ACCESS.2019.2894676]
[http://dx.doi.org/10.1109/ACCESS.2021.3093005] [PMID: 34476144]
[http://dx.doi.org/10.1109/TNB.2012.2214232] [PMID: 22987127]
[http://dx.doi.org/10.1109/TBME.2019.2897285] [PMID: 30716030]
[http://dx.doi.org/10.1080/14737159.2020.1826312] [PMID: 32954863]
[http://dx.doi.org/10.1093/bfgp/elaa009] [PMID: 32393978]
[http://dx.doi.org/10.1186/s13023-021-01839-9] [PMID: 33962631]
[http://dx.doi.org/10.1101/2020.12.20.20248572]
[PMID: 36477409]
[http://dx.doi.org/10.1038/s41564-022-01233-6] [PMID: 36266338]
[http://dx.doi.org/10.1186/s13073-022-01026-w] [PMID: 35220969]
[http://dx.doi.org/10.1002/ajmg.a.63062] [PMID: 36563179]
[http://dx.doi.org/10.1111/tbed.14749] [PMID: 36316791]
[http://dx.doi.org/10.23919/IConAC.2018.8748972]
[http://dx.doi.org/10.1109/ICICCSP53532.2022.9862476]
[http://dx.doi.org/10.1109/ICDABI51230.2020.9325640]
[http://dx.doi.org/10.1007/s11517-022-02591-3] [PMID: 35668230]
[http://dx.doi.org/10.1002/ctm2.28] [PMID: 32508008]
[http://dx.doi.org/10.1109/ICTCS.2019.8923053]
[http://dx.doi.org/10.1007/s00521-022-07121-8]
[http://dx.doi.org/10.1007/s13204-021-01868-7]
[http://dx.doi.org/10.1016/j.bspc.2022.104192] [PMID: 36168586]
 25
25 3
3