Abstract
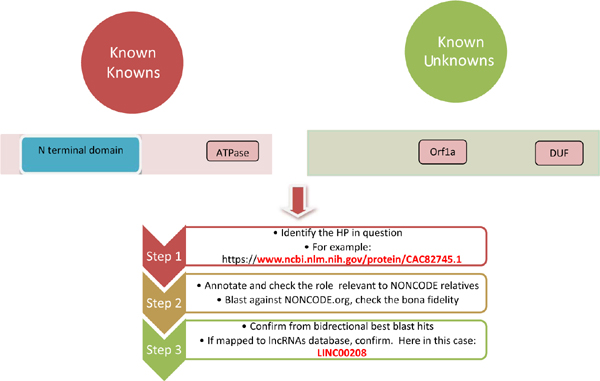
Hypothetical Proteins [HP] are the transcripts predicted to be expressed in an organism, but no evidence of it exists in gene banks. On the other hand, long non-coding RNAs [lncRNAs] are the transcripts that might be present in the 5’ UTR or intergenic regions of the genes whose lengths are above 200 bases. With the known unknown [KU] regions in the genomes rapidly existing in gene banks, there is a need to understand the role of open reading frames in the context of annotation. In this commentary, we emphasize that HPs could indeed be the predecessors of lncRNAs.
Keywords: Hypothetical proteins, lncRNA, aptamers, annotation, functional genomics, transcripts.
[http://dx.doi.org/10.1093/jxb/erp043] [PMID: 19269994]
[http://dx.doi.org/10.1111/j.1574-6968.2001.tb10814.x]
[http://dx.doi.org/10.1016/S0958-1669(99)00063-4]
[http://dx.doi.org/10.4103/ijmy.ijmy_111_17] [PMID: 29171451]
[http://dx.doi.org/10.1016/j.str.2008.10.017] [PMID: 19081051]
[http://dx.doi.org/10.1371/journal.pone.0084263] [PMID: 24391926]
[http://dx.doi.org/10.1093/gigascience/giy150] [PMID: 30520990]
[http://dx.doi.org/10.1021/acs.jproteome.8b00262] [PMID: 29757649]
[http://dx.doi.org/10.1073/pnas.0509109102] [PMID: 16332960]
[http://dx.doi.org/10.1007/s11693-014-9148-4] [PMID: 24799963]
[http://dx.doi.org/10.1109/BIBM.2012.6392719]
[http://dx.doi.org/10.2174/0929866525666180828110444] [PMID: 30152276]
[http://dx.doi.org/10.1038/520615a]
[http://dx.doi.org/10.1186/s12859-018-2554-y] [PMID: 30621574]
[http://dx.doi.org/10.1504/IJBRA.2014.065247] [PMID: 25335568]
[http://dx.doi.org/10.1093/nar/gks915]
[http://dx.doi.org/10.12688/f1000research.9118.1] [PMID: 27853512]
[http://dx.doi.org/10.1002/jcc.25522] [PMID: 30368845]
[http://dx.doi.org/10.1186/s12859-017-1723-8] [PMID: 28655296]
[http://dx.doi.org/10.1109/PAAP.2011.36]
[http://dx.doi.org/10.3390/ijms20112845] [PMID: 31212665]
[http://dx.doi.org/10.1038/s41598-019-43708-3] [PMID: 31089211]
[http://dx.doi.org/10.1007/s13361-015-1161-7] [PMID: 26002791]
[http://dx.doi.org/10.1007/978-1-60761-977-2_12]
[http://dx.doi.org/10.3390/s120100612] [PMID: 22368488]
 53
53