Abstract
Background: Celery (Apium graveolens L.) is considered one of the most important cash crops of Apiaceae family. However, due to minimal genetic and genomic resources, most of the past studies were based on physiological and biochemical analysis rather than molecular breeding.
Objective: The present in silico molecular study is an attempt to develop dbEST derived simple sequence repeats in celery.
Method: Implementation of computational tools and analysis for the development and identification of EST-SSR markers of celery. Developed microsatellite markers were verified using biotechnological techniques.
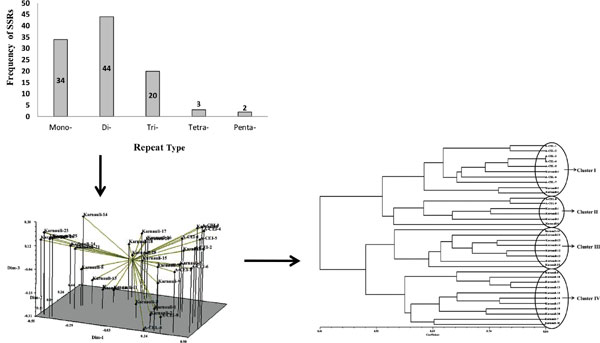
Results: We derived 118 SSRs, of which 103 (87.3%) were simple and 15 SSRs (12.7%) were compound in nature. Of the various SSRs 34 (33%) were mononucleotide, 44 (43%) dinucleotide, 20 (19%) trinucleotide, 3 (3%) tetranucleotide and 2 were penta-nucleotides. Genetic diversity and relationship among 35 local cultivars of celery were identified. Experiments were also conducted to investigate the competence of primers generated for celery genotypes.
Conclusion: Thirty nine microsatellite makers were recognized best for distinguishing celery genotypes. This study provides a scientific ground for celery genotype identification and differentiation, evolutionary genetics, phylogenetics studies, breeding and protection of breeders’ rights on celery as well as on different Apiaceae species in future.
Keywords: Apiaceae, celery, genotypes, in silico, seed spices, markers, microsatellite.
 41
41 3
3