Abstract
Background: Tremor is one of the most noticeable features, which occurs during the early stages of Parkinson’s Disease (PD). It is one of the major pathological hallmarks and does not have any interpreted mechanism. In this study, we have framed a hypothesis and deciphered protein- protein interactions between the proteins involved in impairment in sodium and calcium ion channels and thus cause synaptic plasticity leading to a tremor.
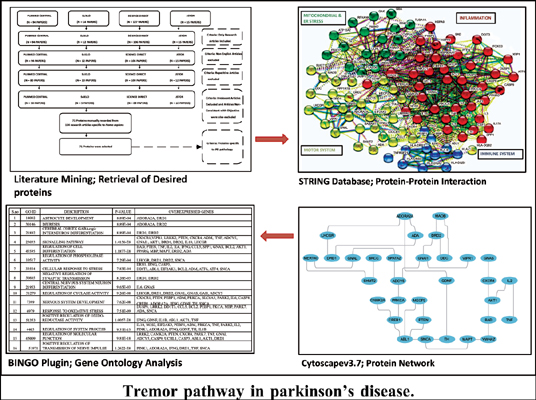
Methods: Literature mining for retrieval of proteins was done using Science Direct, PubMed Central, SciELO and JSTOR databases. A well-thought approach was used, and a list of differentially expressed proteins in PD was collected from different sources. A total of 71 proteins were retrieved, and a protein interaction network was constructed between them by using Cytoscape.v.3.7. The network was further analysed using the BiNGO plugin for retrieval of overrepresented biological processes in Tremor-PD datasets. Hub nodes were also generated in the network.
Results: The Tremor-PD pathway was deciphered, which demonstrates the cascade of protein interactions that might lead to tremors in PD. Major proteins involved were LRRK2, TUBA1A, TRAF6, HSPA5, ADORA2A, DRD1, DRD2, SNCA, ADCY5, TH, etc.
Conclusion: In the current study, it is predicted that ADORA2A and DRD1/DRD2 are equally contributing to the progression of the disease by inhibiting the activity of adenylyl cyclase and thereby increases the permeability of the blood-brain barrier, causing an influx of neurotransmitters and together they alter the level of dopamine in the brain which eventually leads to tremor.
Keywords: Parkinson's disease, tremors, cytoscape, interaction analysis, bingo tool, protein dysregulation.
[PMID: 22033559]
[http://dx.doi.org/10.4103/0976-3147.176192]
[http://dx.doi.org/10.1016/j.cmet.2012.08.012] [PMID: 23168220]
[http://dx.doi.org/10.1155/2016/9832839]
[http://dx.doi.org/10.1093/brain/aws023] [PMID: 22382359]
[http://dx.doi.org/10.1016/j.jns.2016.04.041] [PMID: 27288772]
[http://dx.doi.org/10.1586/14737175.2014.877842] [PMID: 24471711]
[http://dx.doi.org/10.4161/auto.5.5.8505] [PMID: 19377297]
[http://dx.doi.org/10.1016/j.neurobiolaging.2008.11.008] [PMID: 19329225]
[http://dx.doi.org/10.1002/ana.20256] [PMID: 15349860]
[http://dx.doi.org/10.1242/dmm.028738] [PMID: 28468938]
[http://dx.doi.org/10.32607/20758251-2015-7-4-146-149] [PMID: 26798503]
[http://dx.doi.org/10.1038/srep24475] [PMID: 27080380]
[http://dx.doi.org/10.1111/ane.12181] [PMID: 24032478]
[http://dx.doi.org/10.3389/fneur.2018.00109] [PMID: 29541058]
[http://dx.doi.org/10.1016/j.parkreldis.2015.11.016] [PMID: 26627941]
[http://dx.doi.org/10.1016/j.gendis.2016.04.004] [PMID: 30258892]
[http://dx.doi.org/10.1016/j.neulet.2013.10.060] [PMID: 24211691]
[http://dx.doi.org/10.1186/s12864-017-4098-3] [PMID: 28899360]
[http://dx.doi.org/10.1002/mds.26598] [PMID: 27061943]
[http://dx.doi.org/10.1074/jbc.M113.457408] [PMID: 23532841]
[PMID: 29201184]
[http://dx.doi.org/10.1093/bioinformatics/bti551] [PMID: 15972284]
[http://dx.doi.org/10.3389/fnmol.2016.00089] [PMID: 27708561]
[http://dx.doi.org/10.1038/nrn2038] [PMID: 17180163]
[http://dx.doi.org/10.1007/s12640-009-9076-3] [PMID: 19551455]
[http://dx.doi.org/10.1155/2015/628192]
[http://dx.doi.org/10.1007/s10571-011-9671-8] [PMID: 21547489]
[http://dx.doi.org/10.1002/mds.23851] [PMID: 21812034]
[http://dx.doi.org/10.1002/1873-3468.12989] [PMID: 29364506]
[http://dx.doi.org/10.3390/ijms17060954] [PMID: 27314344]
[http://dx.doi.org/10.1093/hmg/ddt530] [PMID: 24158851]
[http://dx.doi.org/10.1523/JNEUROSCI.21-07-02247.2001] [PMID: 11264300]
[http://dx.doi.org/10.1016/j.cellsig.2014.08.019] [PMID: 25173700]
[http://dx.doi.org/10.1016/0304-3940(96)12706-3] [PMID: 8809836]
[PMID: 26261796]
[http://dx.doi.org/10.3389/fnagi.2015.00039] [PMID: 25870559]
[http://dx.doi.org/10.1016/S1474-4422(10)70254-4] [PMID: 20970382]
[http://dx.doi.org/10.1007/s11064-011-0637-5] [PMID: 22083667]
[http://dx.doi.org/10.1038/s41598-019-55479-y] [PMID: 31827228]
[http://dx.doi.org/10.2217/pgs.15.23] [PMID: 25872644]
[http://dx.doi.org/10.3390/molecules22040676] [PMID: 28441750]
[http://dx.doi.org/10.1007/s00702-018-1903-3] [PMID: 29995172]
[http://dx.doi.org/10.1371/journal.pone.0160925] [PMID: 27508417]
[http://dx.doi.org/10.1016/j.neuron.2014.12.007] [PMID: 25611507]
[http://dx.doi.org/10.1046/j.1365-2125.2000.00259.x] [PMID: 11012552]
[http://dx.doi.org/10.1093/brain/awh054] [PMID: 14662519]
[http://dx.doi.org/10.1124/pr.110.002642] [PMID: 21303898]
[http://dx.doi.org/10.1523/JNEUROSCI.2590-10.2010] [PMID: 21123574]
[http://dx.doi.org/10.1016/j.procbio.2013.02.030]
[http://dx.doi.org/10.1111/bph.12181] [PMID: 23517078]
[http://dx.doi.org/10.1016/j.ajhg.2011.04.017] [PMID: 21596366]
[http://dx.doi.org/10.1042/BJ20121022] [PMID: 23282092]
[http://dx.doi.org/10.1016/j.jad.2011.10.030] [PMID: 22119081]
[http://dx.doi.org/10.1016/j.nbd.2016.04.008] [PMID: 27151770]
[http://dx.doi.org/10.4161/auto.25239] [PMID: 23778835]
[http://dx.doi.org/10.1038/nrm906] [PMID: 12209126]
[http://dx.doi.org/10.2174/1389202914666131210211033] [PMID: 24532986]
[http://dx.doi.org/10.1080/15548627.2015.1121360] [PMID: 26902584]
[PMID: 19750042]
[http://dx.doi.org/10.1016/j.conb.2003.11.001] [PMID: 14662371]
 74
74